The classic paper on the EM algorithm begins with a little application in multinomial modeling:
Rao (1965, pp. 368-369) presents data in which 197 animals are distributed multinomially into four categories, so that the observed data consist of 
A genetic model for the population specifies cell probabilities  for some
for some  with
with  .
.
Solving this analytically sounds very much like a 1960’s pastime to me (answer:  , and a modern computer with PyMC can make short work of numerically approximating this maximum likelihood estimation. It takes no more than this:
, and a modern computer with PyMC can make short work of numerically approximating this maximum likelihood estimation. It takes no more than this:
import pymc as mc, numpy as np
pi = mc.Uniform('pi', 0, 1)
y = mc.Multinomial('y', n=197,
p=[.5 + .25*pi, .25*(1-pi), .25*(1-pi), .25*pi],
value=[125, 18, 20, 34], observed=True)
mc.MAP([pi,y]).fit()
print pi.value
But the point is to warm-up, not to innovate. The EM way is to introduce a latent variable  and set
and set  , and work with a multinomial distribution for
, and work with a multinomial distribution for  that induces the multinomial distribution above on
that induces the multinomial distribution above on 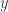 . The art is determining a good latent variable, mapping, and distribution, and “good” is meant in terms of the computation it yields:
. The art is determining a good latent variable, mapping, and distribution, and “good” is meant in terms of the computation it yields:
import pymc as mc, numpy as np
pi = mc.Uniform('pi', 0, 1)
@mc.stochastic
def x(n=197, p=[.5, .25*pi, .25*(1-pi), .25*(1-pi), .25*pi], value=[125.,0.,18.,20.,34.]):
return mc.multinomial_like(np.round_(value), n, p)
@mc.observed
def y(x=x, value=[125, 18, 20, 34]):
if np.allclose([x[0]+x[1], x[2], x[3], x[4]], value):
return 0
else:
return -np.inf
It is no longer possible to get a good fit to an mc.MAP object for this model (why?), but EM does not need to. The EM approach is to alternate between two steps:
- Update
 (E-step, because it is set to its conditional expectation based on the current value of
(E-step, because it is set to its conditional expectation based on the current value of  )
)
- Update
 (M-step, because it is chosen to maximize the likelihood for the current value of
(M-step, because it is chosen to maximize the likelihood for the current value of  )
)
This is simply implemented in PyMC and quick to get the goods:
def E_step():
x.value = [125 * .5 / (.5 + .25*pi.value), 125 * .25*pi.value / (.5 + .25*pi.value), 18, 20, 34]
def M_step():
pi.value = (x.value[1] + 34.) / (x.value[1] + 34. + 18. + 20.)
for i in range(5):
print 'iter %2d: pi=%0.4f, X=%s' %(i, pi.value, x.value)
E_step()
M_step()
It is not fair to compare speeds without making both run to the same tolerance, but when that is added EM is 2.5 times faster. That could be important sometimes, I suppose.

